- Nu.Q® Technology
Our technology detects characteristic epigenetic changes in nucleosomes that occur from the earliest stages of cancer, sepsis and other diseases.
-
- Our Tests
- Human Health
- Nu.Q® NETs
Nu.Q® NETs is a groundbreaking CE-marked diagnostic solution that clinicians can use to detect NETosis.
- Nu.Q® Discover
Buy our Nu.Q® Discover H3.1 Research Use Only Assay
- Animal Health
- Nu.Q® Vet Cancer Test
Nu.Q® Vet Cancer Test is an affordable, accessible blood test that detects cancer in dogs.

Enabling drug developers and scientists access to a range of state-of-the-art assays for rapid epigenetic profiling.
Nu.Q® Discover
Rapid epigenetic profiling for drug developers.
What is Nu.Q® Discover?
Our Nu.Q® Discover program enables drug developers and scientists access to a range of state-of-the-art assays for rapid epigenetic profiling in disease model development, preclinical testing, and clinical studies from discovery to market ready.
Nu.Q® Discover is built on our proprietary nucleosome quantification technology. It is an invaluable research tool for R&D professionals working within the field of pharmacoepigenetics, studying the epigenetic basis for variation in response to drugs.

Labs around the world using Nu.Q® Discover
Assays available
You can use our assays to answer your clinical questions, such as measuring treatment efficacy, or on-target and off-target effects in drug development.
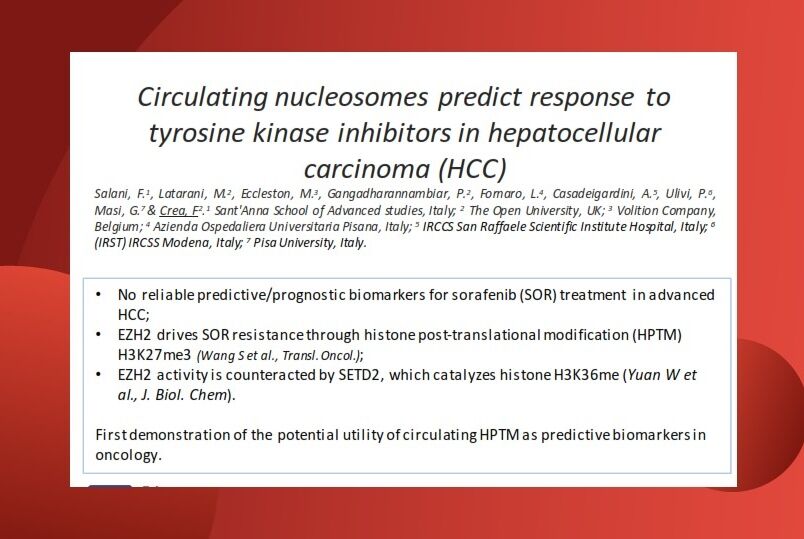
The role of nucleosomes and H3.1
Nucleosomes are small fragments of chromosomes released into the blood during cell death and consist of a histone octamer core with DNA wrapped around it. Histone proteins could be modified by a variety of post-translational modifications (PTMs) also called epigenetic modifications, such as methylation, acetylation, phosphorylation or citrullination. In epigenetics, modifications to histones, including H3, play a key role in regulating gene expression.
Measuring and monitoring
Measuring and monitoring nucleosome levels and modifications in circulating blood has the potential to aid diagnosis, prognosis and monitoring of many human diseases.
Nu.Q® Discover is built on Volition’s cutting-edge nucleosome quantification technology and adds value across the drug development pipeline, with applications in:
- Oncology
- Neurodegenerative diseases
- Autoimmune diseases
- NETosis
Our biomarkers
Our biomarkers support the entire drug discovery and development process from pre-clinical testing to market-ready. We aim to assess disease severity, monitor treatment response, and enhance the understanding of disease pathology and treatments.


Who can work with us?
If you are a pharmaceutical company or an academic research institution, you can access our range of state-of-the-art assays to answer your clinical questions, such as measuring treatment efficacy or on-target and off-target effects in drug development.
If you are a drug developer or scientist you can either:
- Buy our Nu.Q® Discover H3.1 Research Use Only Assay directly.
- Or work with us, access our assays and realize your longer term, drug development needs.
Chief Innovation Officer, Volition
Brochure & Clinical Papers
Contact Nu.Q® Discover Team
Modal Contact Form (Discover)
"*" indicates required fields