- Nu.Q® Technology
Our technology detects characteristic epigenetic changes in nucleosomes that occur from the earliest stages of cancer, sepsis and other diseases.
-
- Our Tests
- Human Health
- Nu.Q® NETs
Nu.Q® NETs is a groundbreaking CE-marked diagnostic solution that clinicians can use to detect NETosis.
- Nu.Q® Discover
Buy our Nu.Q® Discover H3.1 Research Use Only Assay
- Animal Health
- Nu.Q® Vet Cancer Test
Nu.Q® Vet Cancer Test is an affordable, accessible blood test that detects cancer in dogs.



Work with us.
Work with us to access our range of proprietary, state-of-the-art assays and answer your drug development, pre-clinical and clinical questions.
Nu.Q® Discover
How we can help.
Nu.Q® Discover is a complete solution to profiling nucleosomes and empowers drug developers and scientists offering rapid epigenetic profiling in disease model development, preclinical testing, and clinical studies – from drug discovery to market ready.
Nu.Q® Discover is an invaluable research tool for R&D professionals working within the field of Pharmacoepigenetics, studying the epigenetic basis for variation in response to drugs.
Applications
Nu.Q® Discover is built on Volition’s cutting-edge Nu.Q® nucleosome quantification technology and adds value across the drug development pipeline, with applications in:
- Oncology
- Neurodegenerative diseases
- Autoimmune diseases
- NETosis
Use our Nu.Q® Discover assays to test sensitively for circulating nucleosomes and gain deep insights into how drug actions are impacted by epigenetic alterations.
Our biomarkers support the entire drug discovery and development process from pre-clinical testing to market ready.
-
Identify potential drug targets by measuring levels of specific histone modifications.
-
Screen and identify lead compounds that modulate the levels of specific histone modifications.
-
Monitor changes in histone modification levels to assess the efficacy and Mechanism of Action (MoA) of compounds.
-
Monitor target engagement to determine the pharmacodynamics (PD) of drugs in patients.
-
Demonstrate dose/time response in disease/treatment monitoring.
Identify
- Drug targets/novel inhibitors
- Mechanism of Action (MoA)
- Biomarkers
Demonstrate
- Pharmacodynamics (PD) monitoring of target engagement
- On/Off target activities and effects
- Correlation with gene expression and cancer progression
- Efficacy: in-vitro/in-vivo
Accelerate
- Use same assay for drug screen on cell culture to clinical studies
- Demonstrated in a variety of cells and animal models
- Combine assays for a comprehensive overview
Explore our assays
Our range of Nu.Q® Discover assays are for Research Use Only and not for use in diagnostic procedures.

H3.1
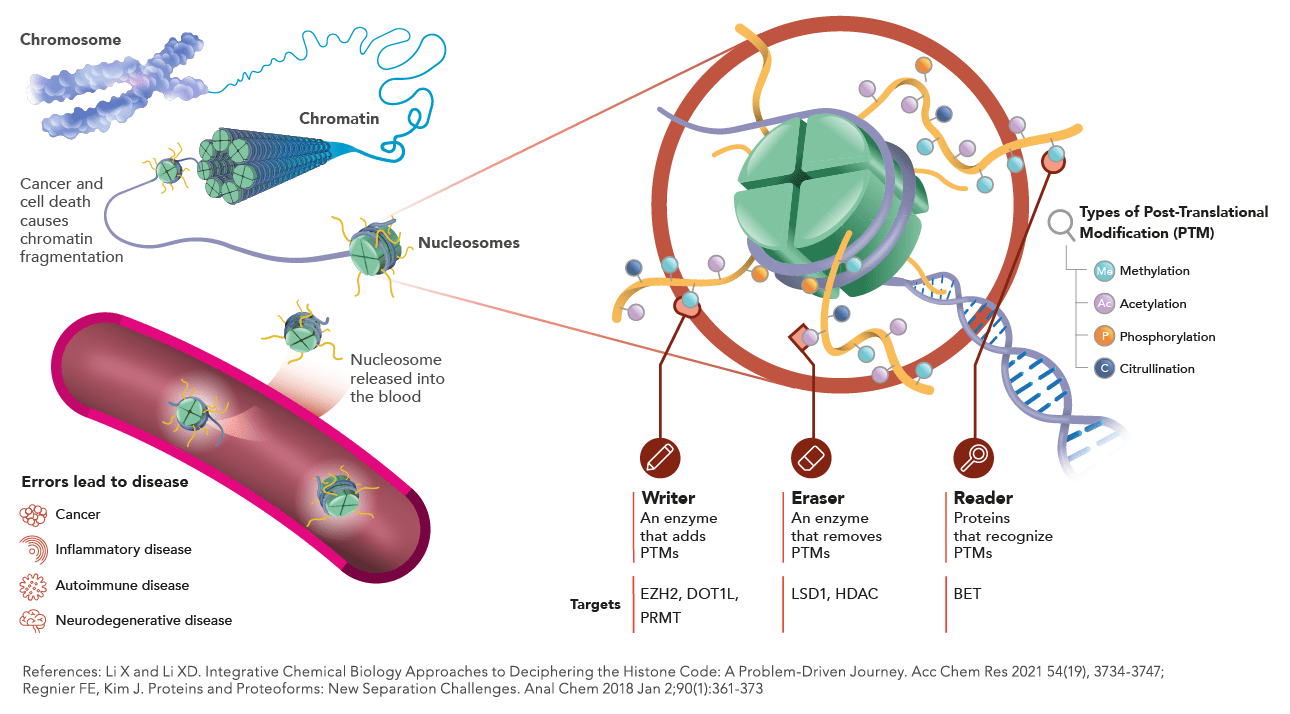
The nucleosome core particle is composed of an octamer of histone proteins comprising a tetramer of histone H3 and H4 and two dimers H2A-H2B, around which 147bp of DNA is wrapped. In addition to the post-translational modifications occurring on the N-terminal tails of the histone, histone variants (for histone H3, H2A and H2B but not H4) could be incorporated. The canonical H3.1 histone variant, incorporated during S-phase, is maintained at high levels in cells dividing at a high rate but is massively evicted in cells undergoing their last cell cycle before exit to differentiation.

H3K27Me3
Epigenetic modification on the Histone 3 (H3). This epigenetic mark indicates the tri-methylation of lysine 27 on H3 protein. This modification is mainly associated with genes repression and often found in heterochromatin regions. H3K27Me3 is regulated by Enhancer of Zeste (EZH2) of Polycomb Repressive Complex PRC2. H3K27me3 has been implicated in numerous of diseases including cancer and it has also been linked to the repair of DNA damage.

H3K36Me3
Epigenetic modification on the Histone 3 (H3). This epigenetic mark indicates the tri-methylation of lysine 36 on H3 protein. This modification is often found in gene body of transcriptionally active genes. The trimethylation is mainly catalyzed by SETD2 methyltransferase.

H3K4Me2
Epigenetic modification on the Histone 3 (H3). This mark indicates the di-methylation of lysine 4 on H3 protein and is enriched in cis-regulatory regions in particular, promoters of transcriptionally active genes.

H3K9Me3
Epigenetic modification on the Histone 3 (H3). This epigenetic mark indicates the tri-methylation of lysine 9 on H3 protein and is associated with gene repression and silent hetero- chromatin. H3K9me3 plays a major role in the cell integrity, genomic stability, and heterochromatin maintenance.

H3K9Me1
Epigenetic modification on the Histone 3 (H3). This epigenetic mark indicates the (mono-) methylation of lysine 9 on H3 protein. H3K9Me1 is a critical marker for transcriptional silencing. This mark is a target of HP1 proteins and primes the formation of functional heterochromatin, a critical chromatin landmark for genome stability.

H3K4Me1
Epigenetic modification on the Histone 3 (H3). This epigenetic mark indicates the (mono-) methylation of lysine 4 on H3 protein. Histone H3 lysine 4 (H3K4) methylation is widely regarded as a mark of transcriptional activation. In particular, H3K4Me1 is commonly considered a chromatin hallmark of enhancers (while high level of tri-methylation of lysine 4 on the histone H3 -H3K4Me3- predominantly mark promoters). However, not all H3K4Me1-enriched regions correspond to enhancers, this mark is also present at promoters. This implies that H3K4me1 modification has a context-dependent role in regulating transcription.

H3K18Ac
Epigenetic modification on the Histone 3 (H3). This epigenetic mark indicates the acetylation of lysine 18 on H3 protein. Histone acetylation is commonly linked to active transcription. Indeed, such modifications lead to chromatin relaxation that enables the activation of gene transcription. Moreover, this modification can serve as docking sites for proteins involved in gene activation, such as transcriptional activators and chromatin remodeling complexes.

H3K9Ac
Epigenetic modification on the Histone 3 (H3). This mark indicates the acetylation of lysine 9 on H3 protein. This mark, as H3K14Ac, is linked to gene activation and active promoters.

H3K27Ac
Epigenetic modification on the Histone 3 (H3). This mark indicates the acetylation of lysine 27 on H3 protein. This mark is associated with the higher activation of transcription, and primarily defined as an active enhancer mark. H3K27Ac is also found surrounding transcriptional start sites (TSS).

H3S10Ph
Epigenetic modification of Nucleosome on the Histone 3 (H3). This mark indicates the phosphorylation of serine 10 on H3 protein. This modification, done by Aurora kinase, plays a role in mitosis.

pH2AX
Epigenetic modification on the isoform X of Histone 2A (H2AX). This mark indicates the phosphorylation of serine 139 on H2AX protein. This modification correlates with double-strand DNA damage.

H3R8Cit
Epigenetic modification on the Histone 3 (H3). This mark indicates the citrullination of arginine 8 on H3 protein. Protein arginine deiminase 4 (PAD4) mediates citrullination of histone tail residues.

H3K27M
Mutation in the Histone 3 (H3) on the lysine 27 replaced by a Methionine and is found in aggressive pediatric gliomas.

Case study
Our assays in action
Client: ORIC Pharmaceuticals
Project: Pharmacodynamics (PD) biomarker
We worked with ORIC Pharmaceuticals and demonstrated how the Nu.Q® Discover cell-free (cf)-nucleosomal H3K27me3 assay enables robust, quantitative analysis of histone modifications in circulating cf-nucleosomes.
Findings:
- A method of choice for quantification of plasma cf-nucleosome H3K27me3 levels.
- The assay has shown small molecule inhibition of PRC2 in a manner that is:
- Dose dependent
- Time dependent
- Tumor selective
- The assay is non-invasive and allows for serial sample collection from patients.
- No requirements for further optimization by the investigator / sponsor of the trial.
- Once samples have arrived at Volition, the turnaround time for results is fast < 1 week.
Volition Science Advisory Board Member

Working with us
Get in touch with our commercial team today to discuss your needs.
Our expert team will be on hand to offer guidance and support and to fulfil your needs.
"*" indicates required fields
Get in touch
Want to buy our H3.1 kit?
Our Nu.Q® Discover H3.1 Research Use Only Assay detects quantitative levels of circulating intact nucleosomes containing histone H3 in plasma.
A valuable research tool to support your drug discovery and development needs.
Brochure & Clinical Papers
Contact Nu.Q® Discover Team
Modal Contact Form (Discover Work With Us)
"*" indicates required fields