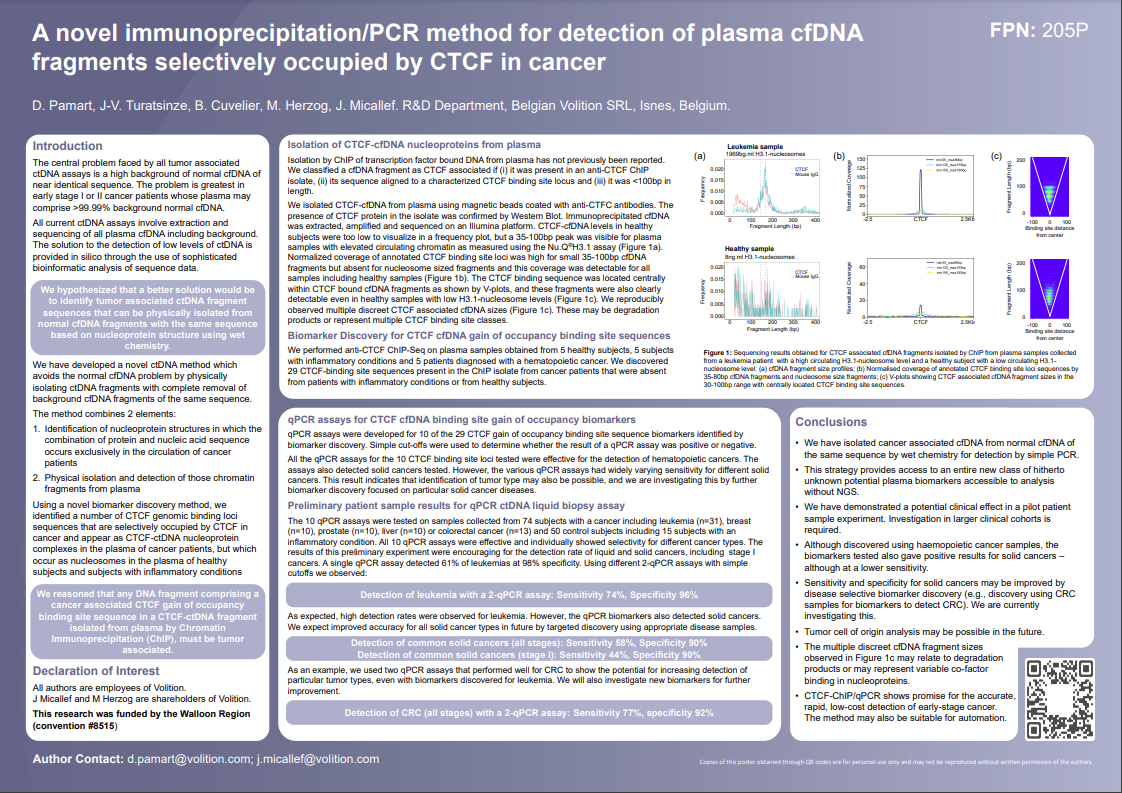
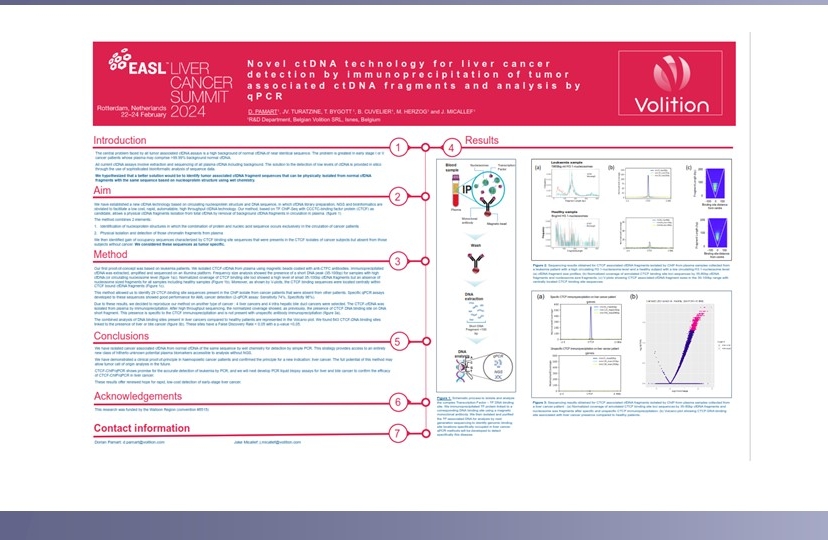
-
Nu.Q® Technology
Our technology detects characteristic epigenetic changes in nucleosomes that occur from the earliest stages of cancer, sepsis and other diseases.
-
- Our Tests
- Human Health
-
Nu.Q® NETs
Nu.Q® NETs is a groundbreaking CE-marked diagnostic solution that clinicians can use to detect NETosis.
-
Nu.Q® Discover
Buy our Nu.Q® Discover H3.1 Research Use Only Assay
- Animal Health
-
Nu.Q® Vet Cancer Test
Nu.Q® Vet Cancer Test detects 76% of systemic cancers at 97% specificity.

Breakthrough Cancer Detection Method.
Capture-PCR™
Early Cancer Detection
In early-stage cancer, it is difficult to detect cancer-derived circulating tumor DNA (ctDNA) in the blood because it may comprise only 0.01% of the DNA present among a background of 99.99% normal DNA. Moreover, most of the cancer DNA has exactly the same sequence as normal DNA.
Current ctDNA detection methods involve DNA extraction, sequencing of all (cancer and normal) circulating DNA and analysis of the sequencing data using sophisticated computer bioinformatics to tell them apart. Physical separation of tumor-derived and healthy circulating DNA has previously never been reported

We have developed Capture-PCR™ – a novel method for liquid biopsy involving the first reported physical isolation of a class of tumor-derived ctDNA fragments from blood.
Cancer-derived ctDNA fragments are then extracted after removal of all normal background DNA of the same sequence for detection with a simple, low cost PCR test.
Research
Read our latest research paper or watch our webinar for more in-depth information.
Capture-PCR™ obviates expensive, time-consuming DNA sequencing and bioinformatics – allowing for rapid, cost-effective detection in a routine blood test. It may also be suitable for automation, enabling application in hospital laboratories.

Fast.
Cost-effective.
Routine blood test.
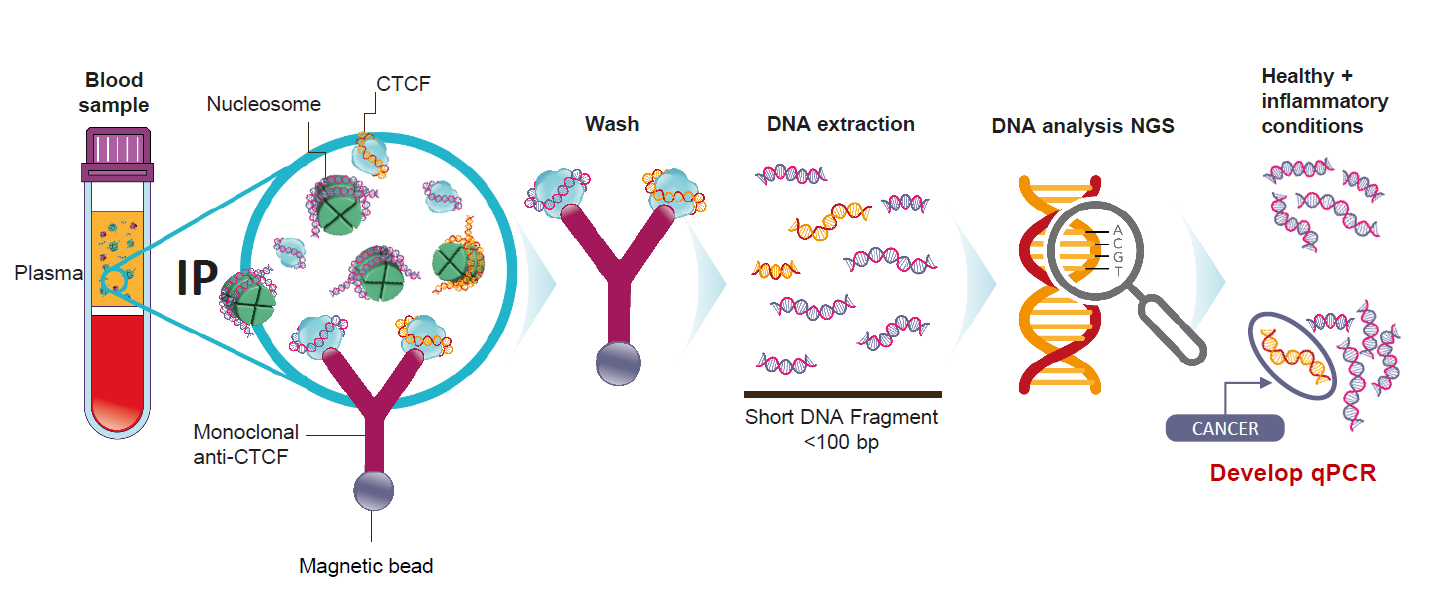
Workflow
Based on over a decade of work on the chemistry of circulating chromatin fragments, we have developed a transformational wet chemistry pathway that identifies and physically isolates chromatin fragments that we know are tumor-derived from background DNA of the same sequence, using Chromatin Immunoprecipitation (ChIP).
Quantitative real-time PCR (qPCR) testing is undertaken to establish whether cancer is present.
Our latest research
We tested Capture-PCR™ in a first small clinical experiment and detected a range of liquid and other cancers, including at early-stage I disease.
- 74% of leukemias were detected at 96% specificity with a single q-PCR assay
- 77% of colorectal cancers were detected at 92% specificity with 2-qPCR assays.
These early assays were developed using a leukemia model, but surprisingly also detected many other cancers including detecting colorectal cancer in a blood test with an accuracy approaching that of current Fecal Immunochemical Tests (FIT).
of leukemias were detected at 96% specificity with a single q-PCR assay
of colorectal cancers were detected at 92% specificity with 2-qPCR assays.
Next Steps
The opportunity.
Capture-PCR™ results to date are exciting and may pave the way for a whole new class of undiscovered biomarkers.
We are now developing a range of cancer-specific assays which we expect to be more accurate and look forward to sharing our progress throughout 2024 and beyond.
Our Capture-PCR™ commercial strategy is to license this proprietary technology to an industry leader/leaders. Contact us for more information.
Professor of Translational Genomics at the Institute of Cancer Research, and Scientific Director of Clinical Genomics at The Royal Marsden NHS Foundation Trust
Brochure & Clinical Papers
Contact us about Breakthrough Cancer Detection
Modal Contact Form (Breakthrough Cancer Detection)
"*" indicates required fields